Want to save time and money getting your NGS tests up and running? Our analytical validation (AV) consulting services can help! Have an AV specialist contact you.
Want to save time and money getting your NGS tests up and running? Our analytical validation (AV) consulting services can help! Have an AV specialist contact you.(availability and levels of service may vary by region)
The human genome is heavily populated with repeat regions which makes designing primers difficult, a well known challenge in PCR design. The Ion AmpliSeq Designer pipeline has been developed to deliver the most robust set of amplicons it can generate. The pipeline specifically excludes amplicons that are placed in repeat elements or other hypervariable regions in order to generate the best possible outcome for actual amplicon coverage when actually used in a reaction.
A focus in our Research & Development department is to better understand the properties of repeat regions to allow primer placement in these regions to achieve higher target design rate while maintaining coverage uniformity and on-target rates.
The Biological filtering mechanism that is incorporated into the Ion AmpliSeq Designer pipeline to evaluate these repeat elements is the RepeatMasker. RepeatMasker is a program that screens DNA sequences for interspersed repeats and low complexity DNA sequences and is an annotation track that is available through the UCSC Genome Browser. One of the new feature upgrades in v1.2 of the Ion AmpliSeq Custom Designer links directly to the browser and offers the user the visual representation to distinguish between three BED files as custom annotation tracks.
While G-C bonds contribute more to the stability, and hence increased melting temperature, of the primer/template binding than do A-T bonds, it is important to keep in mind that two primer / template complexes with similar or even identical content of GC may result in a completely different melting temperature because of base order influences towards the overall stability.
GC-rich regions for the target DNA are difficult to amplify, and are generally avoided when defining an e-silico algorithm.
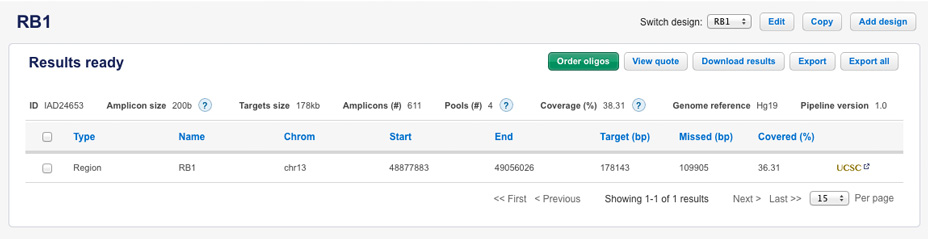
When submitted as a manual target region with coordinates: chr13:48877883-49056026, the resulting coverage was 38.31% with 4 pools and 611 amplicons.
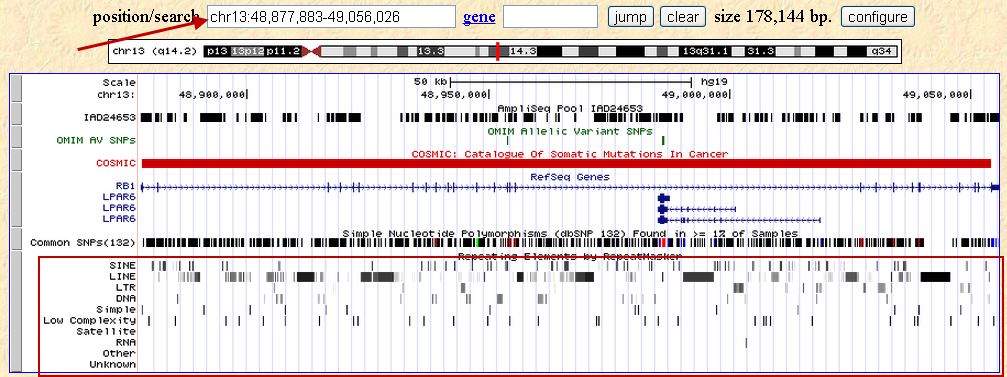
In closer examination of the design results, it is apparent that the majority of this region is interspersed with repeat elements.
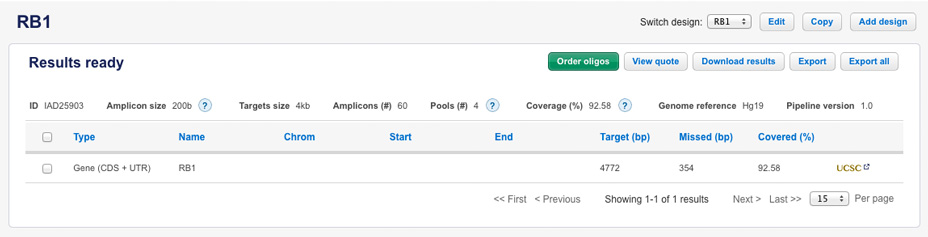
An attempt to re-design the region by input type Gene + UTR results in coverage of 92.58% with 4 pools and 60 amplicons.